InGene Module - Data Analysis
The InGene module creates a data analysis environment aimed at identifying clusters of patients characterized by specific correlations between genetic data and phenotype (and clinical data in general).
The development of the InGene module was aimed at creating an environment in which the operator, be it the geneticist or the data analyst, can be able to define a dataset of genetic data possibly filtered through constraints linked to clinical data and/or instrumental.
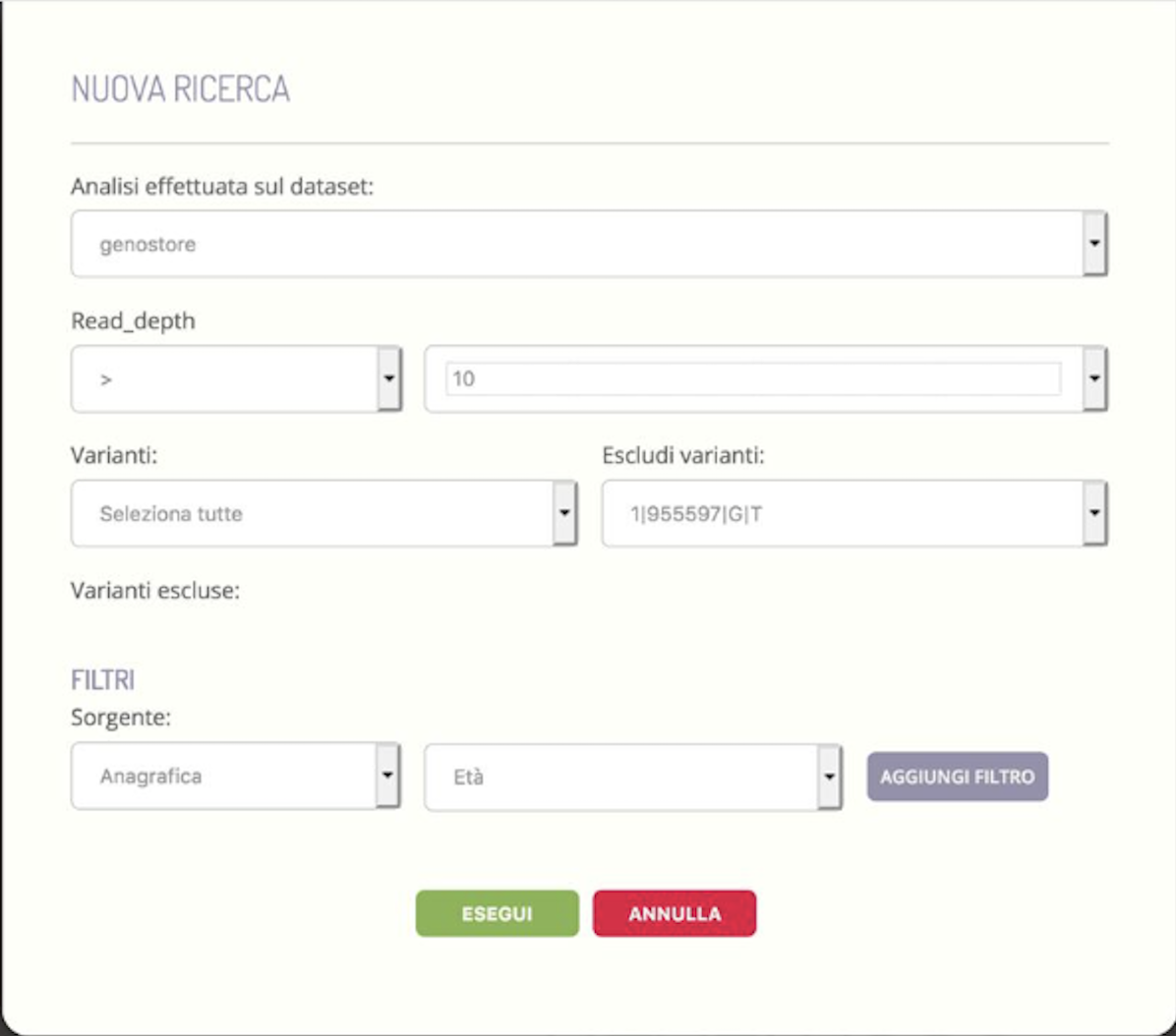
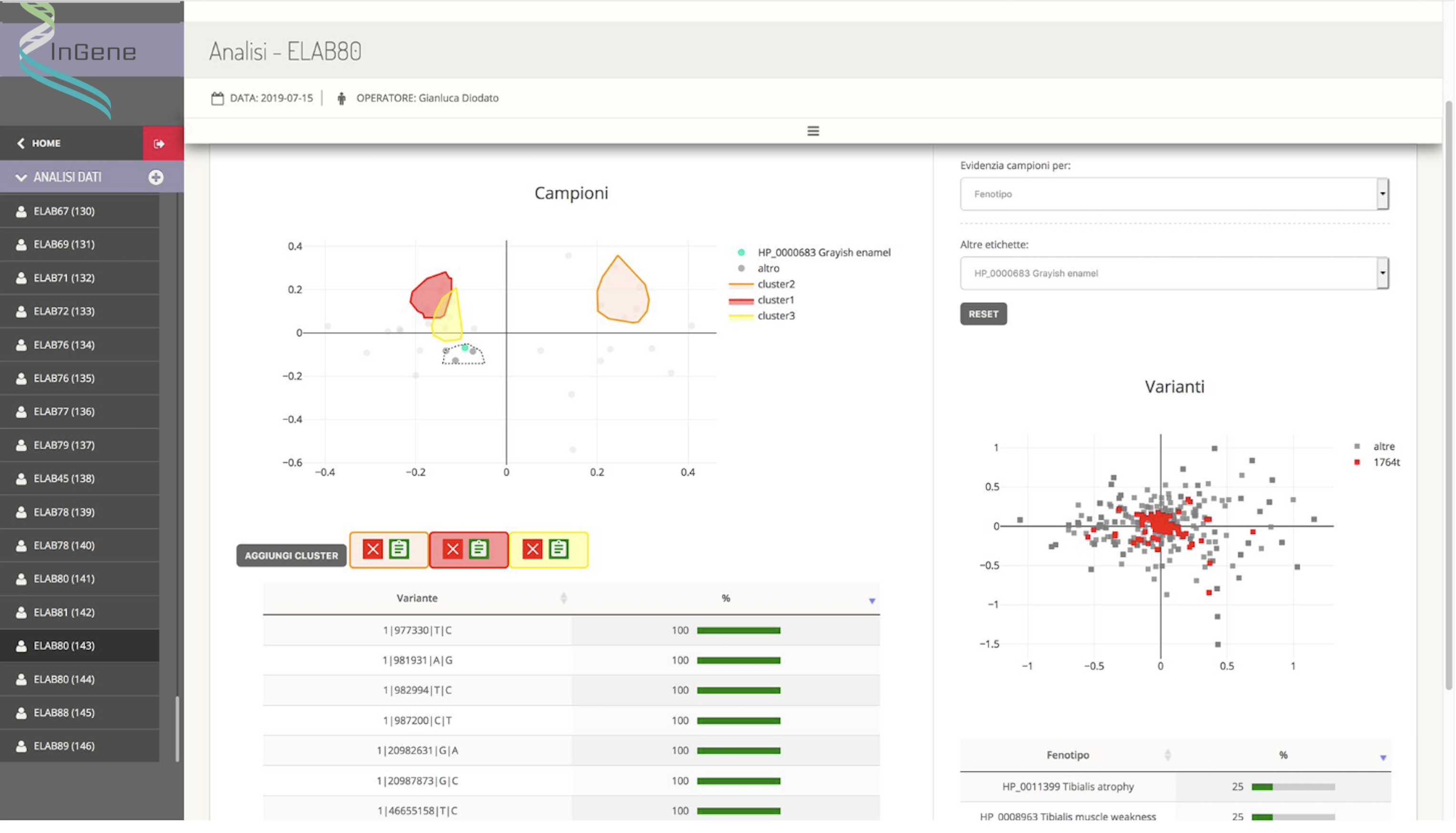
The appropriately defined dataset is then processed by an external module (created by the sub-contractor Kode s.r.l., having a key role in this activity) which applies multivariate statistical analysis techniques that allow the differences and similarities between the samples and between the variations. Specifically, samples that are graphically close to each other appear to have a genetic background similar to each other, while variants that are graphically close to each other are identified in sets of samples that are similar to each other.
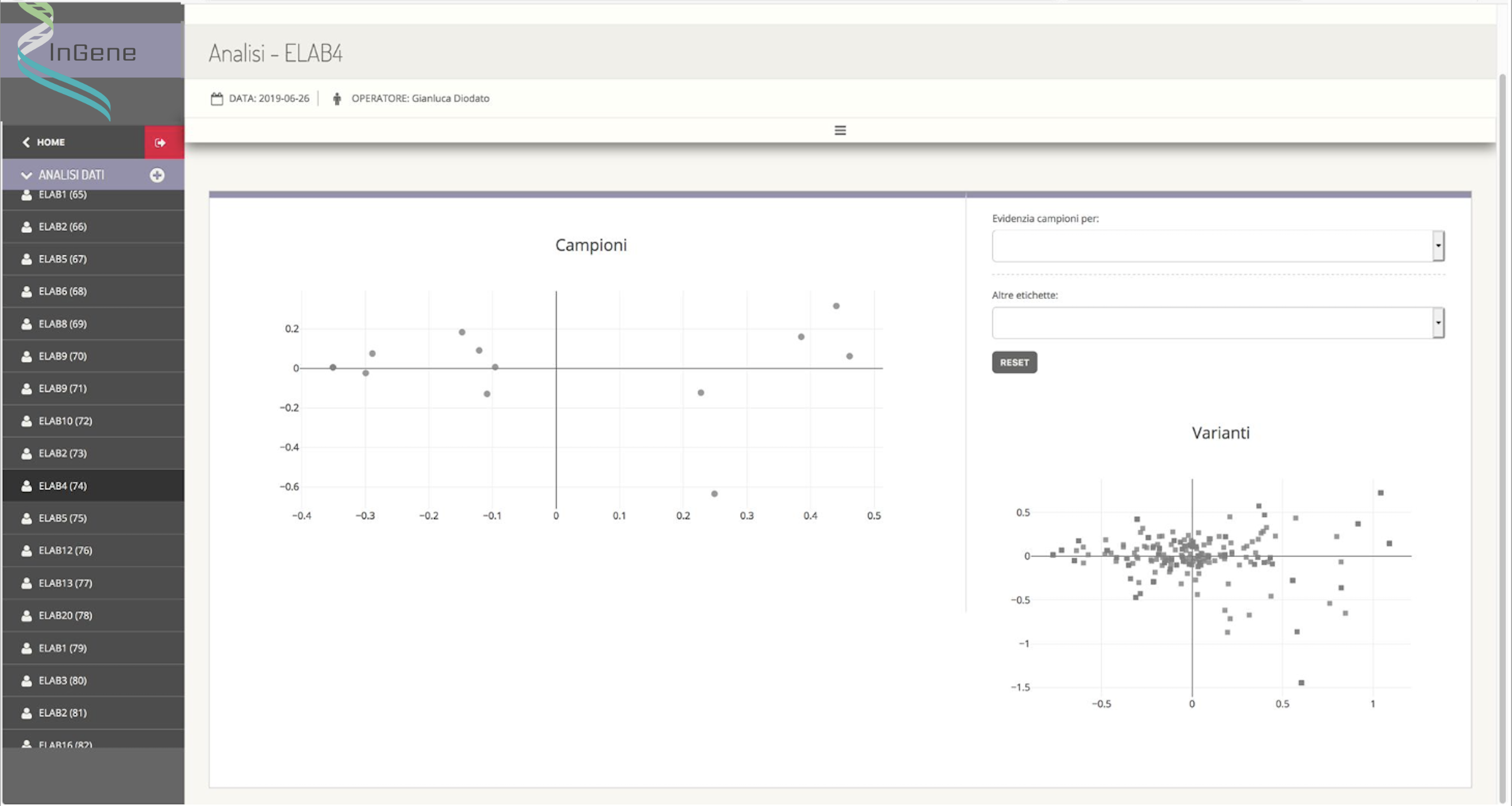
InGene therefore, from the graphical representation of similarities between samples and between variants, aims to:
- superimpose and correlate the distribution of subjects with respect to their clinical data with that relating to their genetic background;
- identify clusters of samples with real-time indication of:
- incidence of individual genetic variants, in terms of percentage rate of presence of a variant in the cluster samples compared to the number of the cluster;
- distribution in terms of percentage rate of presence of the phenotypic characteristics in the population identified by the cluster. Furthermore, for each cluster, all statistical information relating to clinical and genetic data is accessible and can be selected to compose a printable pdf report.